Shift eDNA - Assessing species distribution changes using eDNA
Marine ecosystems are currently suffering from oceanic warming due to global climate change. Fish communities are particularly threatened as most of fish species life cycle is directly influenced by temperatures (e.g. growth, reproduction, recruitment). As such, rise in oceanic temperatures is likely to lead to different changes in species geographic distribution. Some species could colonize and settle in ecosystems at higher latitudes thanks to temperatures becoming more suitable while others could disappear from their initial ecological niche due to too high temperatures, especially at low latitudes.

The “Shift eDNA” project aims to implement a long-term monitoring in different types of ecosystems worldwide (polar, temperate, tropical) to assess species range shift, based on eDNA metabarcoding. This non-invasive technique allows to detect species occurrence based on DNA traces left in the environment, with the advantage of being able to detect small, rare, or crypto-benthic species often missed by traditional sampling methods. Therefore, the use of eDNA metabarcoding is relevant to build an eDNA occurrence database from which we will investigate potential species colonization and/or extirpation in different areas from the tropics to the poles. Moreover, we will be able to model fish species range shift thanks to this database and thus to improve future range shift predictions.
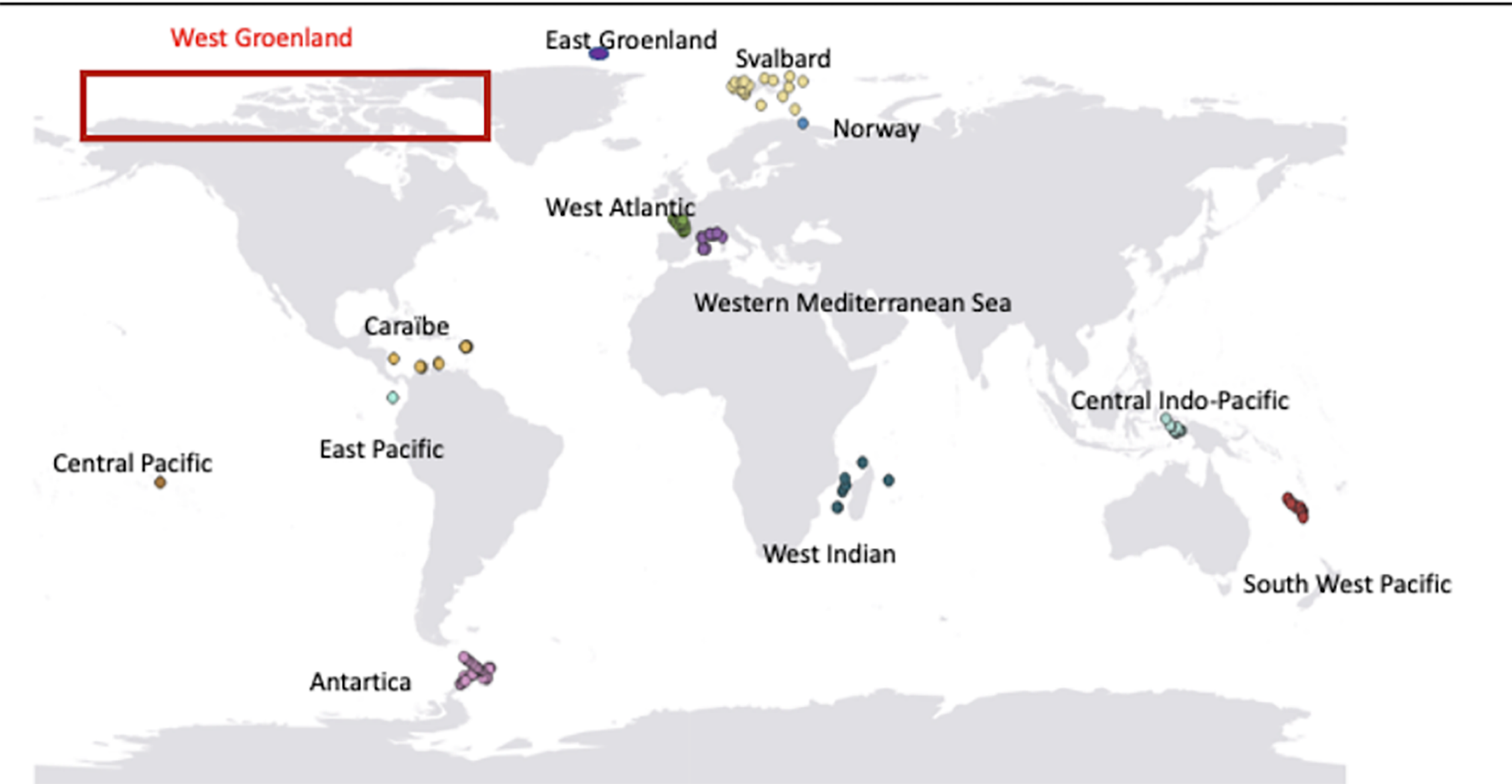
The project is divided into four main parts. The first one corresponds to the eDNA sampling of different areas worldwide. Among all these areas, the Arctic Ocean is of major importance as it has been poorly sampled with traditional methods and is currently experiencing some of the highest warming rates influencing polar biodiversity. These samplings will be combined with the collection of different environmental variables required to build the predictive Species Distribution Models (SDM) and make future projections. In parallel, we will extract fish species occurrence from OBIS (Ocean Biodiversity Information System) to build species distribution model comparable with the eDNA data.
The second part consists in completing the reference genetic database thanks to the sampling and sequencing of the 12S DNA region for a list of missing species (previously determined with GAPeDNA). This step will be based on opportunistic samplings and collaboration with aquariums and museums (especially for rare fishes). Notably, a specific hybrid-capture protocol will be developed to be able to sequence the DNA from fish museum specimens fixed with formaldehyde.
The third part aims to improve eDNA metabarcoding bioinformatic analyses. Therefore, we will develop a deep neural network able to provide matching probabilities to reads and taxonomic units against the reference database, increasing the computing efficiency compared to classical OTU-clustering bioinformatic pipelines. Moreover, geographic information will be implemented as prior in both classical pipelines and deep learning model to refine taxonomic assignation based on eDNA samples locations coupled with known species geographic ranges. Finally, the last part of the project corresponds to the modelling of fish species range shift over time using SDM applied to occurrence data extracted from OBIS (step 1). We will then compare these estimates with the occurrence eDNA data collected worldwide to assess whether they match current and future SDM predictions. These comparisons will allow to detect any type of species range shift in the different ecosystems. Moreover, we will test for the influence of functional traits and phylogeny on these range shifts, to determine if some species-specific functional traits and/or phylogenetic relationships are more likely to favor colonization or extirpation.
The project is conducted in collaboration with the CEFE institute (Montpellier, France), the MARBEC institute (Montpellier, France) and the Cantonal Museum of Natural Sciences (Lausanne, Switzerland).

Sarah Schmid
